Imageomics

My work is centered around Imageomics, an area of research that uses images and machine-learning algorithms to study biology. More specifically, I am focused on integrating pre-existing biological knowledge and interpretable automated-discovery techniques to find new ways of studying evolutionary morphology.
The conclusions we can draw from our data and our models are dependent on both the ways our data interacts with our models and the manner in which our data was collected to begin with. This complication can make studying morphology very difficult, as the way traits have historically been defined through character construction has been dependent on particular experts asking particular questions, which often may not adequately capture the dynamic processes that actually drive evolution more broadly.
Imageomics provides a possible way to get around this, as neural-networks are often able to pick up on complex patterns that humans (even experts) might miss. By grounding them in biological knowledge and enforcing sparse model outputs that encourage inductive inference, we can potentially bypass the interpretability problems associated with black-box models and make character construction more reproducible and more robust to complex evolutionary dynamics.
Automated Character Construction with Deep Learning
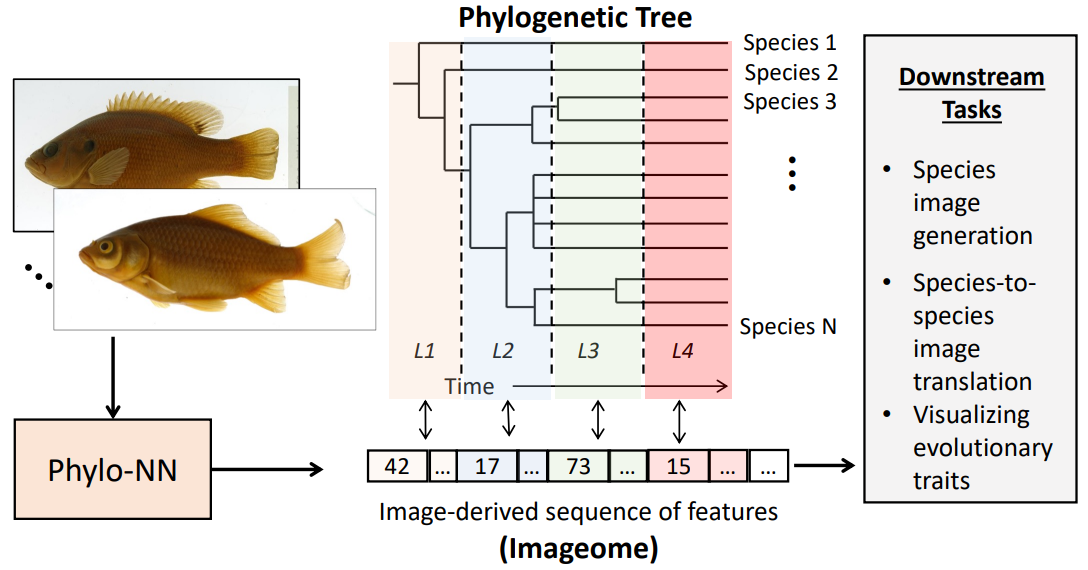
While deep learning models can fit functions to any sort of data, they still always include inductive assumptions about how that data behaves / the processes that led to it being what it is. When dealing with images, convolutional neural networks allow us to incorporate biases about the spatial distribution of patterns. When dealing with time-series data, LSTMs allow us to incorporate biases about the order of elements in a sequence.
When dealing with evolutionary data, we can incorporate similar biases, by using clever restructuring of autoencoders and multi-task learning algorithms to bias our models with information from phylogeny, ontogeny, or other biological processes. In this way, we can ground increasingly high-dimensional, or even raw, datasets in our understanding of reality, and possibily discover new processes that we far more difficult to uncover previously.
Simulating Image Data

Most phenotypic traits are complex characters, whose evolutionary paths are determined by dynamic changes in regulatory networks with various degrees of integration, complexity, and canalization. Deep learning may allow us to uncover these sophisticated processes so that they can be better represented in our models, but many deep learning algorithms are black boxes, and validating the performance of black-box models that uncover unknown processeses in irregularly structured data is incredibly difficult.
One way to address this issue is by instead validating the ability of these models to discover known processes through simulated raw data, which can be done with the TraitBlender pipeline I developed. TraitBlender allows users to represent theoretical morphospaces as python functions, which can be be used to evolve imaginary taxa on a simulated phylogeny. TraitBlender can then take 'museum-specimen' style photos of these imaginary taxa in bulk, allowing users to create huge synthetic image datasets where the evolutionary and developmental processes are known in their entireties.
Incorporating Structured Knowledge into Inference
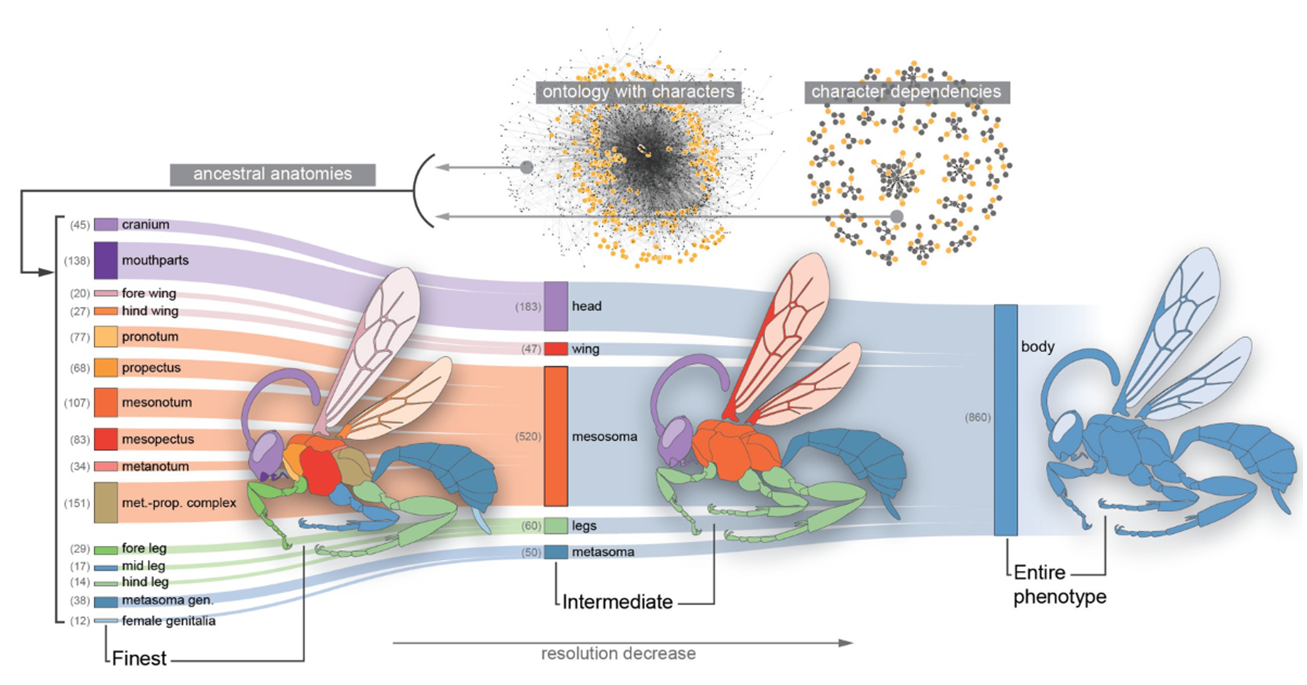
Whenever we make models, we are attempting to codify statements about the world. For a model to be meaningful, however, the assumptions we put into it must be reflective of our assumptions about external reality. When we are dealing with traits that have complex, nonlinear dependencies on each other, doing this can be very difficult.
I've contributed to projects that address this problem by incorporating biological knowledge into models directly in the form of ontologies, which are knowledge graphs that specify the hierarchical relationships / dependencies between traits.
The RPhenoscate Paper | The SCATE (Semantic Comparative Analysis of Trait Evolution) Project